Products You May Like
You share a lot more than just meals and hobbies with your family and friends: you also give each other gut microbes, meaning your personal flora can serve as a detailed profile of your social life.
A new study has found just how much face-to-face social interactions impact the human gut microbiome.
The study, led by Yale University researchers Francesco Beghini and Jackson Pullman, involved pairing a social network map of 1,787 adults living in isolated villages in Honduras with a detailed analysis of the microbes that live in each participant’s gut.
“[It] was a long labor of love (if one can use that expression for the collection of hundreds of stool specimens from isolated jungle villages),” sociologist and physician Nicholas Christakis, also from Yale, told ScienceAlert.
They also collected information on the villagers’ social networks to create a detailed picture of who spent time with who in the community.
The data is drawn from a larger project that began in 2013 in collaboration with local and regional public health agencies and local leaders, and was used not only for research but to provide diagnosis and treatment for participants whose samples reflected a need.
Participants were instructed on how to collect their own stool samples and passed them on to a local team who put them on ice and shipped them off to the US for analysis.
While the larger project involved 176 villages, for this study the team chose to focus on data from 18 particularly isolated villages in the western highlands of Honduras.
“We needed to study isolated populations for our work, where social ties were within a circumscribed community – hence these isolated villages,” Christakis explained.
They plan to conduct similar studies in other parts of the world, like Greece, to see how things compare across different cultures, but Christakis thinks that even this study of remote villages in Honduras offers a universal insight into how human microbiomes are molded by our social structures.
“We believe our findings are of generic relevance, not bound to the specific location we did this work, shedding light on how human social interactions shape the nature and impact of the microbes in our bodies.”
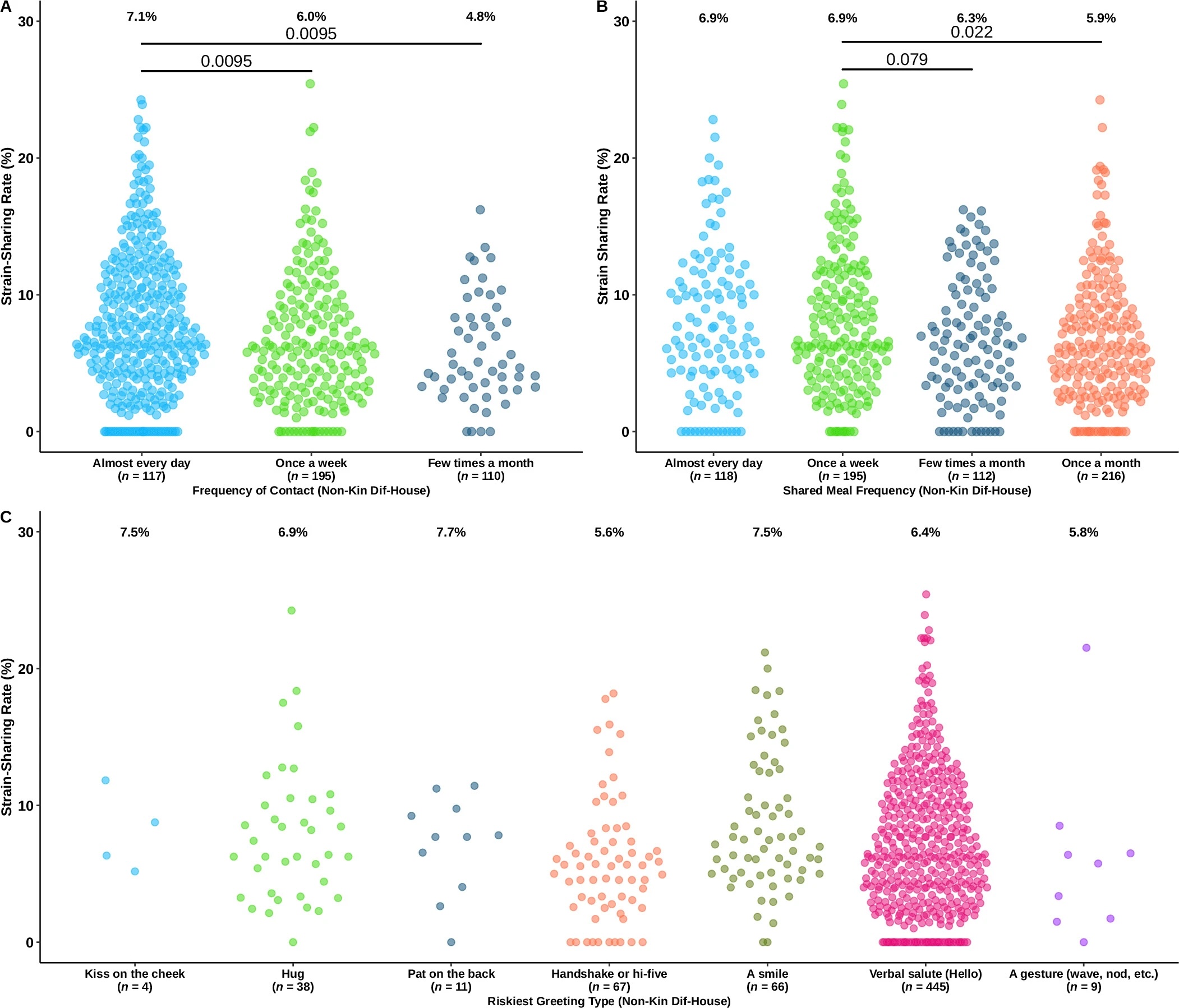
They found that microbial species and strains are shared not only between families, but other non-familial and non-household connections – close friends, for instance.
They also found that the gut flora of socially central people – those who have a greater number of social connections in the community – is more similar to the overall village than people who live on the social periphery.
And that strain-sharing amplifies through social connections over time: among 301 people whose microbiome was measured again after two years, the gut flora strains of those who had more face-to-face connections had become more similar to each other than among otherwise similar co-villagers who were less socially connected.
For those who find themselves increasingly isolated from face-to-face interactions, reduced contact with others is almost certain to play a role in their microbiome’s makeup.
“If you are physically and hence socially isolated, you have different microbes than if you are a social butterfly,” Christakis explained.
But we don’t yet know whether that’s for better or worse. As with most biological phenomena, it probably depends on many factors.
“The sharing of microbes per se is neither good nor bad, but the sharing of particular microbes in particular circumstances can indeed be good or bad,” said Christakis.
“For instance, after a person takes antibiotics, their guts may be denuded of healthy microbes, and they must be recolonized with the healthy, normal microbes we need to function. This recolonization likely often occurs via social interactions.”
Christakis pointed out that studies have linked gut microbiomes with mental and physical health conditions that aren’t otherwise considered biologically contagious, like obesity, depression, and arthritis.
This research suggests that community structure may have an impact on how the microbial profiles of those conditions might emerge.
This research was published in Nature.
